There is overwhelming evidence that our facial features are influenced by our genes.
In most cases, one needn’t look much further than one’s own family photo album to verify this claim.
Nevertheless, very little is known about which genes are most important for determining the shape of a person’s nose or eyes or lips. Uncovering the genetic architecture of such typical facial traits can offer novel insights into the molecular mechanisms that build faces. Moreover, this line of research can enhance our understanding of how craniofacial malformations arise. Down the road, we may even be able to predict someone’s facial features or growth from their DNA, which has numerous potential clinical and biometrics implications.
The first key studies attempting to identify genes that influence quantitative facial traits in humans were published in 2012. These studies and the several that have followed since then (including two from our University of Pittsburgh group) have each identified a handful of potential genes. Some of these signals have been independently replicated – many have not. Such is the case with many complex genetic traits, where we are typically dealing with variants – tiny changes in the DNA – of small effect in relatively small samples.
Now, through a collaborative effort involving research teams at Pitt, KU Leuven (Belgium), Stanford University and Pennsylvania State University, we have achieved a significant advance.
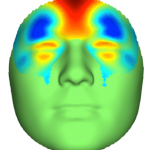
This image shows the effects of a genetic variant at the TBX15 locus on facial morphology as a displacement heat map. Regions in red indicate protrusion, while regions in blue indicate depression. Credit: Claes, et al, Nature Genetics (2018)
The work, published this week in Nature Genetics, has identified several distinct regions in the genome associated with numerous aspects of facial shape. Moreover, we were able to independently and robustly replicate 15 of these genetic signals. The large number of strong genetic associations, coupled with the ability to accurately and intuitively model the effects of genes on faces, make this work stand out.
Many of the genomic regions we identified contained genes that were biologically plausible, either because they have been implicated in known genetic syndromes that affect facial structures or because we know from experimental studies that they are expressed in the developing face. For example, one of our signals implicated the gene TBX15. In our data, we saw a clear association between variants in and around this gene and the protrusion of the forehead and cheekbones. In developing mice, TBX15 is expressed primarily in the frontal portion of the braincase, exactly where one would expect. In humans, mutations in TBX15 result in Cousin syndrome, which is characterized by a prominent bulging forehead. Interestingly, we observed a link between the kind of extreme facial shape traits associated with genetic syndromes and normal-range facial variation again and again. This suggests that while rare damaging mutations in these genes may lead to serious structural birth defects, the more typical variants we observed in our study may have a more benign effect, nudging facial shape within the normal bounds of variability.
One of the keys to our success was the innovative approach to measuring facial traits developed by lead author Dr. Peter Claes, a research scientist in KU Leuven’s Department of Electrical Engineering.
The face – like many parts of the body – is modular in nature, meaning that there are parts or subparts that tend to be highly structurally integrated. We leveraged this native property of faces by using a machine-learning approach to partition the face into distinct modules at multiple overlapping levels of scale, organized hierarchically from the entire head to small localized features.
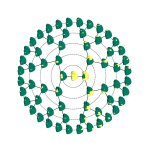
Image showing how the faces in our study were segmented into distinct modules in a hierarchical manner. Modules are in yellow. Credit: Claes, et al, Nature Genetics (2018)
As a result, we have been able to show how different genetic variants relate to facial features in an unprecedented level of detail. Looking at the nose, for example, we observed one set of variants that affected just the tip of the nose, while another set affected only the lateral region where the nostrils are located, while yet another set influenced the nasal root – the area at the top of the nose between the eyes.
An important point is that we did not choose the facial modules beforehand, but instead allowed the modules to emerge in an unsupervised manner from the data. A key feature with this type of data-driven approach to facial measurement is that we are letting the faces tell us which traits are likely to be important, instead of imposing an artificial or arbitrary set of criteria from the outset.
Another critical factor was having access to multiple, high-quality datasets with 3-D facial images and genomic information available. Co-author Dr. Mark Shriver, a professor of anthropology at Penn State, has been collecting facial images and DNA in populations around the world for many years. Our replication cohort was drawn from this wealth of data.
The highly refined genetic associations we uncovered have the potential to provide developmental biologists with a wealth of information – a kind of roadmap – for investigating genes responsible for building faces.
Along these lines, our co-author Dr. Joanna Wysocka, and her team at Stanford University were able to take the genes implicated in this analysis and show that they play a role in regulating the activity of a special population of cells called cranial neural crest cells. Crest cells are responsible for building much of the face during early development. These results are very exciting for us because they are, for the first time, showing us that we may be identifying genetic variants with some potentially important functional effects.
In yet another angle to this story, some of the variants showed marked differences in activity level between chimpanzee and human neural crest cells, suggesting possible evolutionary implications. One variant that we highlight in the paper was located within a known neural crest cell enhancer (a non-coding bit of DNA that can influence how other genes are activated) and showed much greater levels of activity in chimpanzees relative to humans. Interestingly, this particular variant was found to influence the chin region in our study – a uniquely human feature.
But it is easy to get ahead of one’s self. It is important to keep in mind that we are likely just scratching the surface. There are thousands of genes involved in building our faces, and we will need much larger datasets to identify more of these signals. This is our goal over the next few years.
Weinberg is an associate professor at the University of Pittsburgh in the Center for Craniofacial and Dental Genetics (Department of Oral Biology, School of Dental Medicine) with secondary appointments in the Department of Human Genetics and the Department of Anthropology. Other investigators at Pitt involved in this project include Dr. John Shaffer, assistant professor, and Dr. Eleanor Feingold, professor, both in the Department of Human Genetics, Dr. Mary Marazita, professor, and Dr. Jasmien Roosenboom, post-doctoral scholar, both in the Department of Oral Biology.